We offer free open-source tools to support imaging science and foster innovation.
Contact us for training, education, and collaboration.
Image Acquisition, Simulation & Generation
Magnetic Resonance imaging Lab (MRiLab)

MRiLab is a numerical MRI simulation package developed and optimized to simulate MR signal formation, k-space acquisition, and MR image reconstruction. MRiLab simulation platform combined with various toolboxes can be applied to customize virtual MR experiments, which can be a prior stage for prototyping and testing new MR techniques and applications.
Liu F, Velikina JV, Block W, Kijowski R, Samsonov A: Fast Realistic MRI Simulations Based on Generalized Multi-Pool Exchange Tissue Model. IEEE Trans Med Imaging, 2017; 36:527-37.
Generalized Physics-Guided Self-Supervised Learning for RF (GPS-RF)
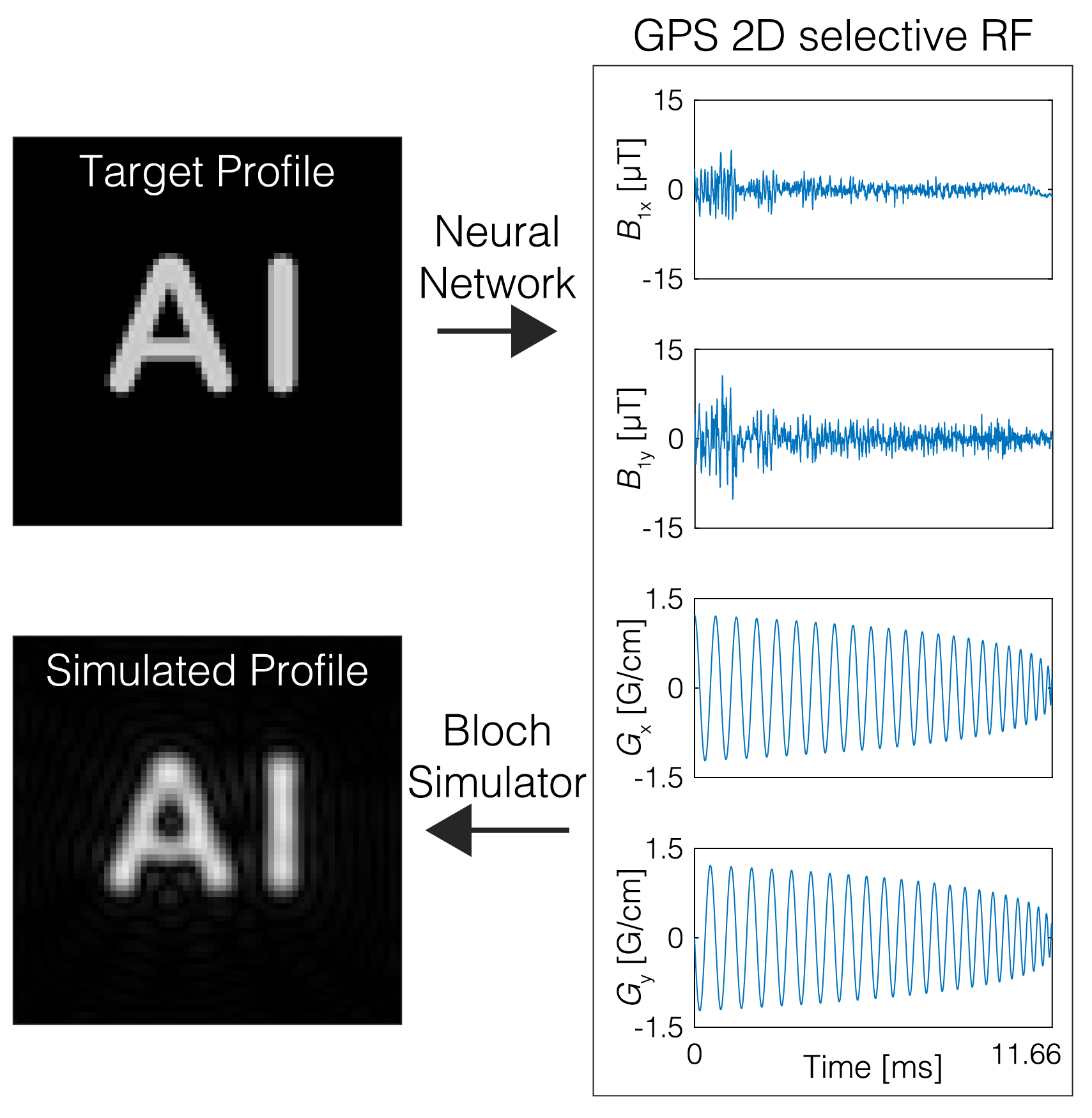
GPS-RF is a comprehensive framework designed for creating MRI RF pulses using a physics-guided, self-supervised learning approach. This algorithm can generate RF pulses that meet flexible design requirements by utilizing a physics module, the Bloch equation, to guide the learning and optimization process.
Jang A, He X, Liu F: Physics-Guided Self-Supervised Learning: Demonstration for Generalized RF Pulse Design. Magn Reson Med. 2024, DOI: 10.1002/mrm.30307
Image Reconstruction
Foundation Model MRI Reconstruction
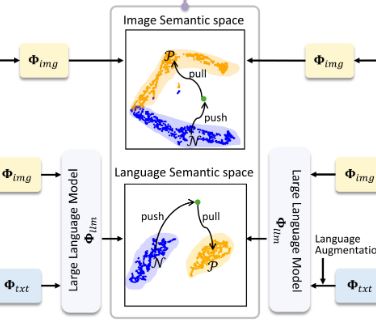
We investigate vision-language foundation models for enhance undersampled MRI reconstruction by leverages high-level semantic embeddings from pretrained vision-language foundation models (specifically Janus) to guide the reconstruction process through contrastive learning. This aligns the reconstructed image embedding with a target semantic distribution, ensuring consistency with high-level perceptual cues.
Feng R, He X, Mercer R, Stewart Z, Liu F: On the Utility of Foundation Models for Fast MRI: Vision-Language-Guided Image Reconstruction. Arxiv Preprint. 2025, DOI: 10.48550/arXiv.2511.19641
Reference-Free Implicit Neural Representation with Model Reinforcement (REFINE-MORE)
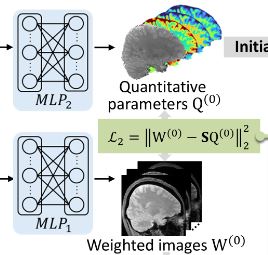
REFINE-MORE is a self-supervised scan-specific method for multiparametric quantitative MRI reconstruction, which integrates the implicit neural representation with MR physics-based model reinforcement.
Feng R, Jang A, He X, Liu F: Accelerating Multiparametric Quantitative MRI Using Self-Supervised Scan-Specific Implicit Neural Representation With Model Reinforcement. Magn Reson Med. 2025, DOI: 10.1002/mrm.70227
POSition Encoding for improved qMRI (POSE)
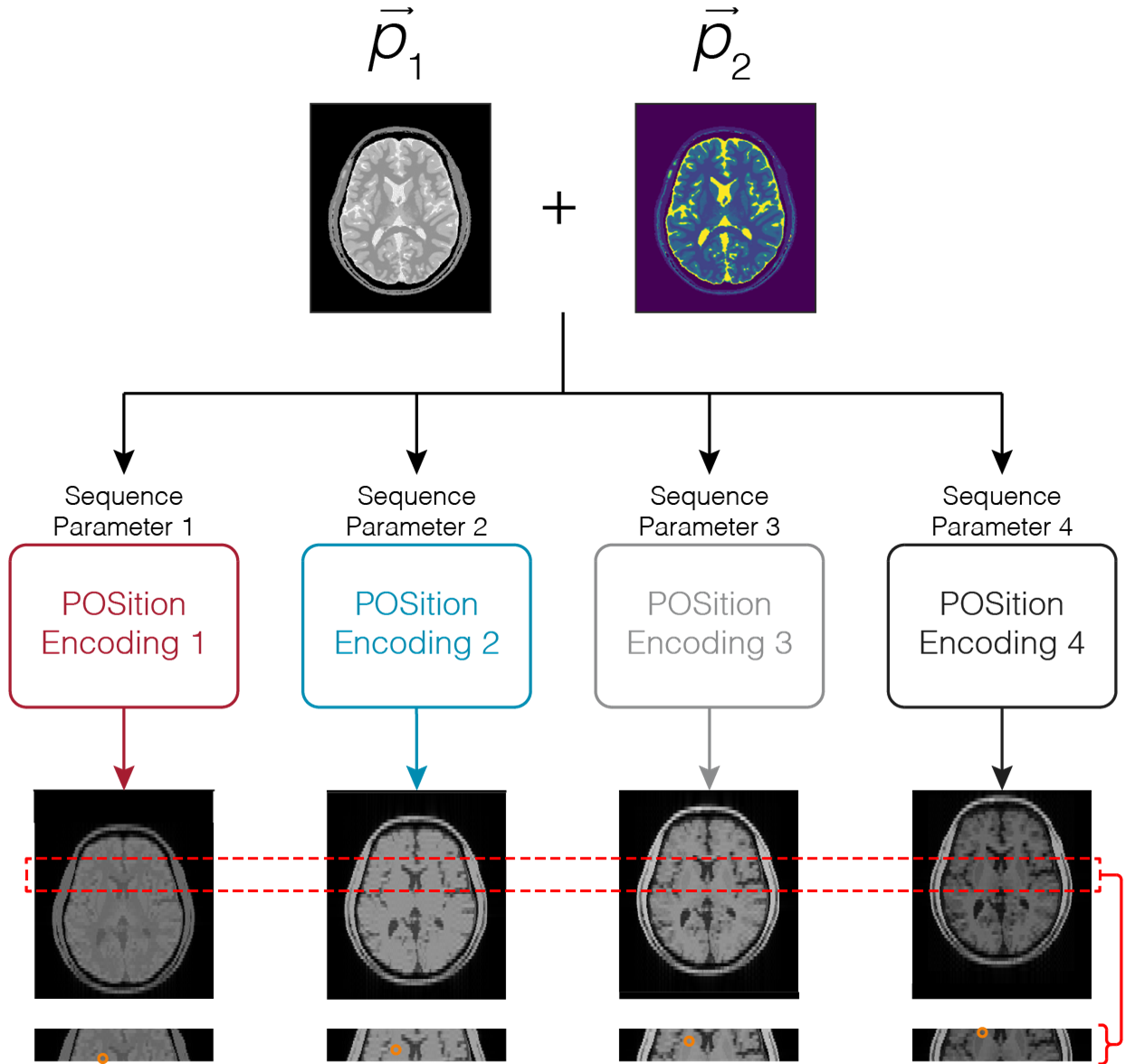
POSE is a technique that uses subvoxel shifting as a source of encoding for accelerating quantitative MRI. The POSE framework applies unique subvoxel shifts along the acquisition parameter dimension, thereby creating an extra source of encoding. Combining with a biophysical signal model of interest, accelerated and enhanced resolution maps of biophysical parameters are obtained. (coming soon)
Jang A, Liu F: POSE: POSition Encoding for accelerated quantitative MRI. Magn Reson Imaging. 2024, DOI: 10.1016/j.mri.2024.110239
Image Analysis & Processing
Matrix User (MatrixUser)
MatrixUser is a software package designed and optimized for manipulating multi-dimensional image data. MatrixUser provides a user-friendly graphical environment for multi-dimensional image display, matrix (image stack) processing, and volume rendering. This lightweight software is a great tool for researchers who use Matlab for image processing and analysis.
Liu F, Samsonov A, Wilson J, Blankenbaker D, Block W, Kijowski R: Rapid In Vivo Multi-Component T2 Mapping of Human Knee Menisci. J Magn Reson Imaging. 2015; 42(5): 1321-8.
Few-shot Adaptation of Training-free SAM (FATE-SAM)
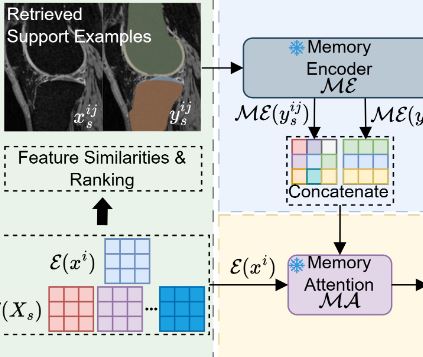
FATE-SAM is a versatile framework for 3D medical image segmentation that adapts the pretrained SAM2 model without fine-tuning. By leveraging a support example-based memory mechanism and volumetric consistency, FATE-SAM enables prompt-free, training-free segmentation across diverse medical datasets.
He X, Hu Y, Zhou Z, Jarraya M, Liu F: Few-Shot Adaptation of Training-Free Foundation Model for 3D Medical Image Segmentation. Arxiv Preprint. 2025, DOI: 10.48550/arXiv.2501.09138