Artificial Intelligence for Medical Imaging
We develop novel AI methods to improve image acquisition and reconstruction, automate image analysis and processing, and enable fully-automated computer-assisted disease diagnosis and prediction.
A full list of our publications in this category can be found here.
Research Highlight
AI and Machine Learning for Rapid MRI
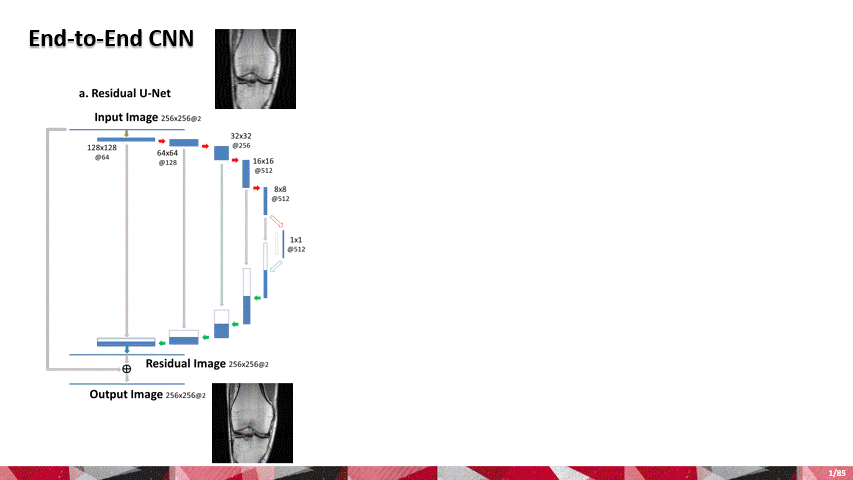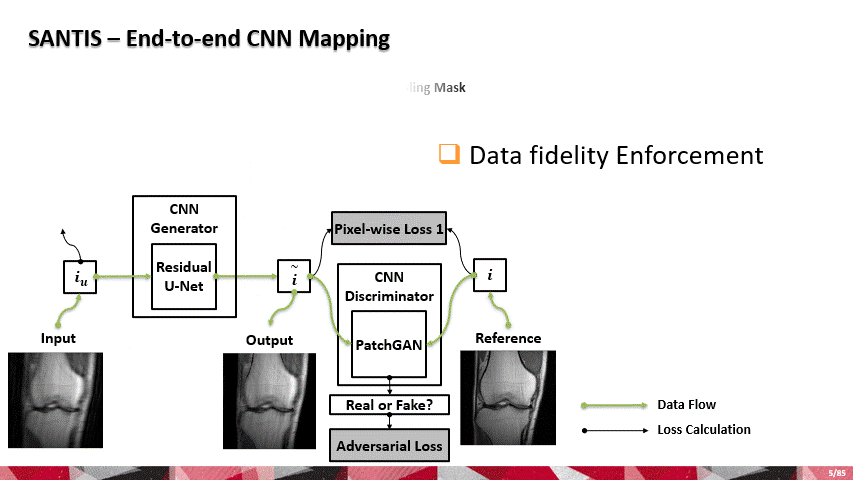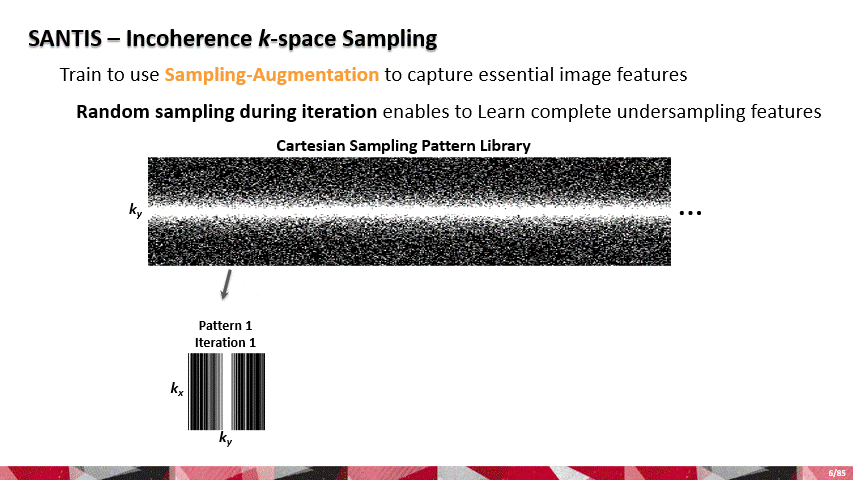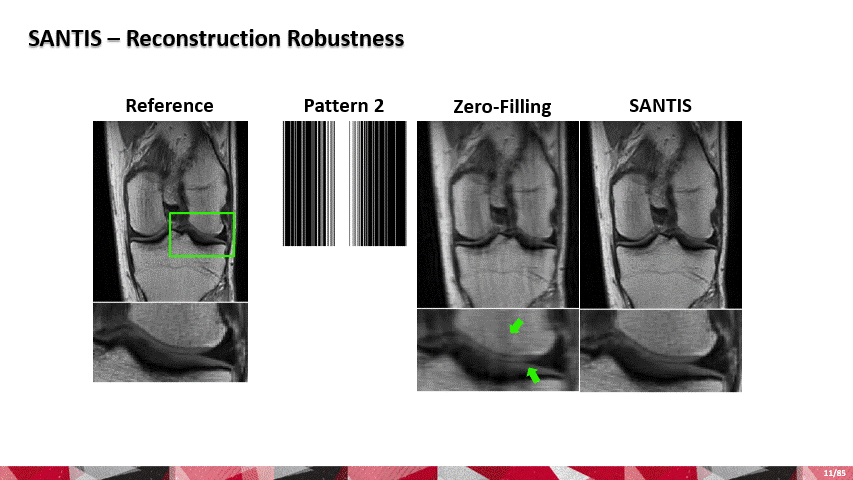
Over the past few years, AI and machine learning have demonstrated the ability to provide improved image quality for reconstructing undersampled MRI data, providing new opportunities to improve the performance of rapid MRI further. Compared to conventional rapid imaging techniques, machine learning-based methods reformulate image reconstruction into a task of feature learning by inferencing undersampled image structures from a large image database. Such data-driven approaches have efficiently removed artifacts from undersampled images, translated k-space information into images, estimated missing k-space data, and reconstructed MR parameter maps. We invented a new method to advance AI-based image reconstruction by rethinking the essential reconstruction components of efficiency, accuracy, and robustness. This new framework, named Sampling-Augmented Neural neTwork with Incoherent Structure (SANTIS), uses a unique recipe of data cycle-consistent adversarial network combining efficient end-to-end convolutional neural network mapping, data fidelity enforcement and adversarial training for reconstructing undersampled MR images.
SANTIS was evaluated to reconstruct undersampled knee images with a Cartesian k-space sampling scheme and undersampled liver images with a non-repeating golden-angle radial sampling scheme. SANTIS demonstrated superior reconstruction performance in both datasets and significantly improved robustness and efficiency compared to several reference methods.
Presentation in ISMRM 27th Annual Meeting Educational Session
AI for Accelerated and Improved Quantitative MRI
Our team invented a novel AI framework for accelerating MR parameter mapping. We demonstrated the AI framework’s efficacy, efficiency, and robustness in resolving the challenges of performing rapid quantitative MR imaging. Among many MR techniques, quantitative mapping of MR parameters has always been shown as a powerful tool for improved assessment of various diseases. In contrast to conventional MRI, parameter mapping can provide increased sensitivity to tissue pathologies with more specific information on tissue composition and microstructure. However, standard approaches for estimating MR parameters usually obtain repeated acquisition of data sets with varying imaging parameters, requiring long scan times. Accelerated methods are highly desirable and remain a hot topic of great interest in the MR community.
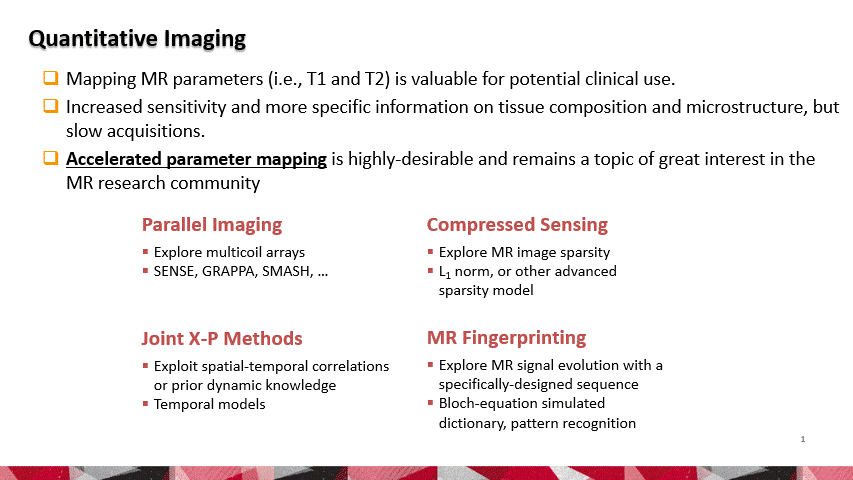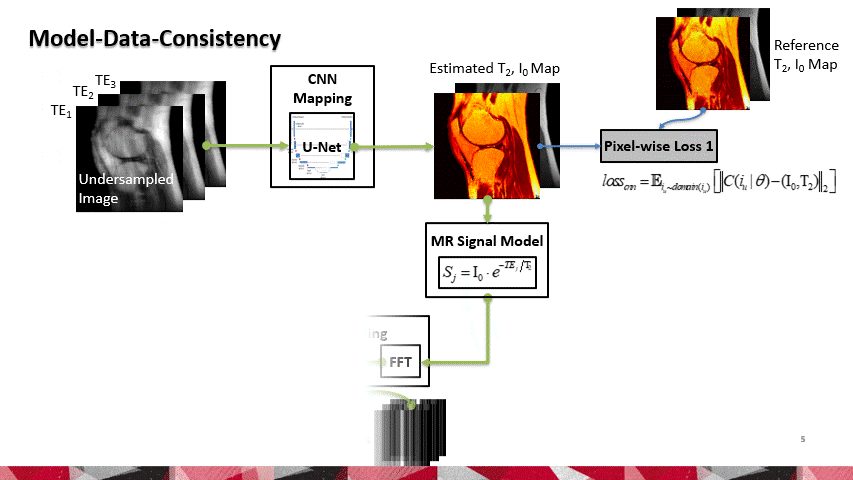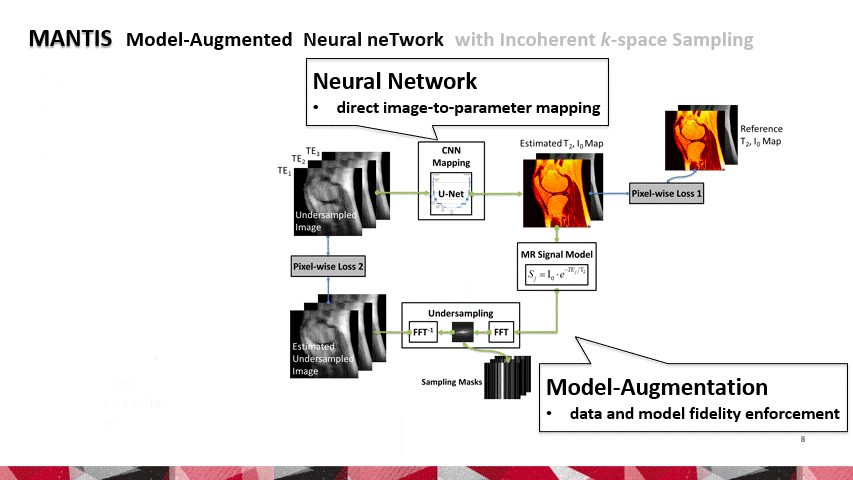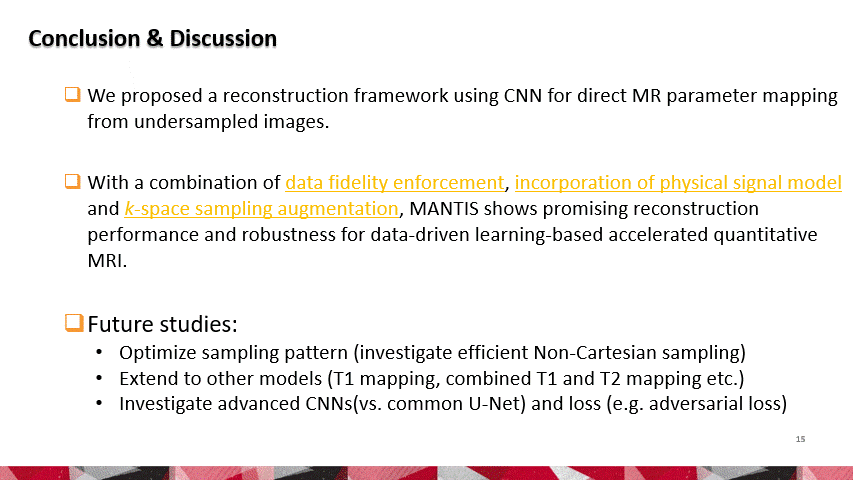
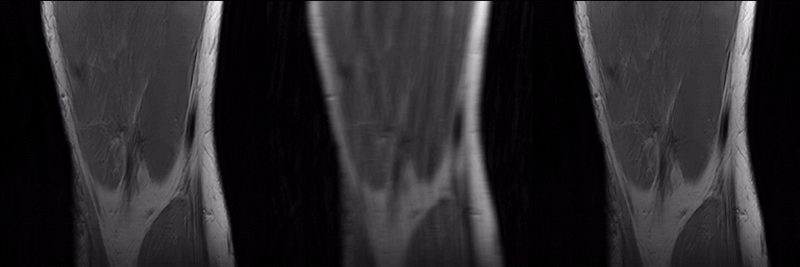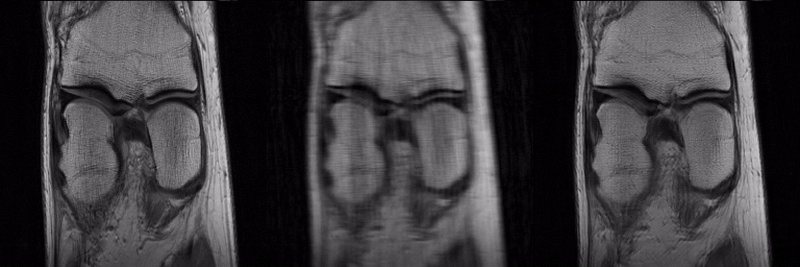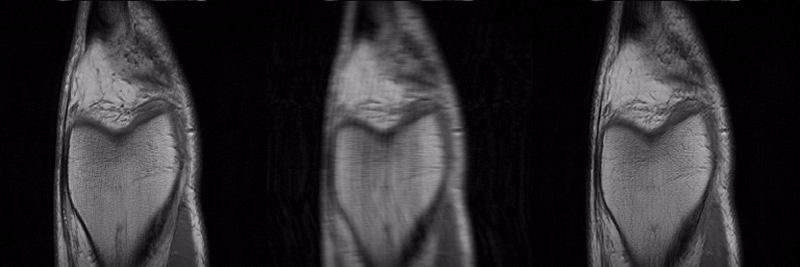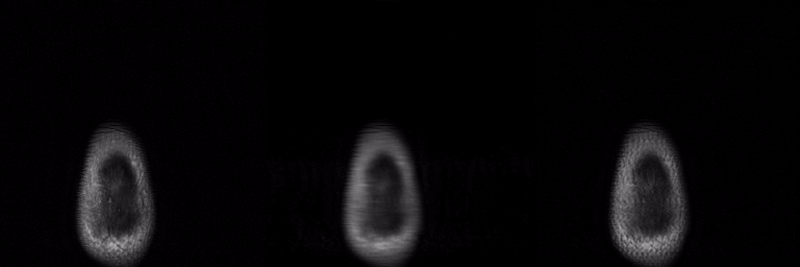
To reconstruct MR parameter maps with less data, we invented MANTIS standing for Model‐Augmented Neural neTwork with Incoherent k‐space Sampling. Our MANTIS algorithm combines end‐to‐end convolutional neural network mapping, model augmentation promoting data consistency, and incoherent k‐space undersampling as a synergistic framework. The CNN mapping converts a series of undersampled images straight into MR parameter maps, representing an efficient cross-domain transform learning. Signal model fidelity is enforced by connecting a pathway between the undersampled k‐space and estimated parameter maps to ensure that the algorithm constructs efficacious parameter maps consistent with the acquired k-space measurements. A randomized k-space undersampling strategy is tailored to create incoherent sampling patterns that are benign to the reconstruction network and adequate to characterize robust image features.
Our study demonstrated that the MANTIS framework represents a promising approach for rapid T2 mapping in knee imaging with up to 8-fold acceleration. In future work, MANTIS can extend to other types of parameter mapping, such as T1 imaging, T1ρ imaging, diffusion, and perfusion, with appropriate models and training data sets.
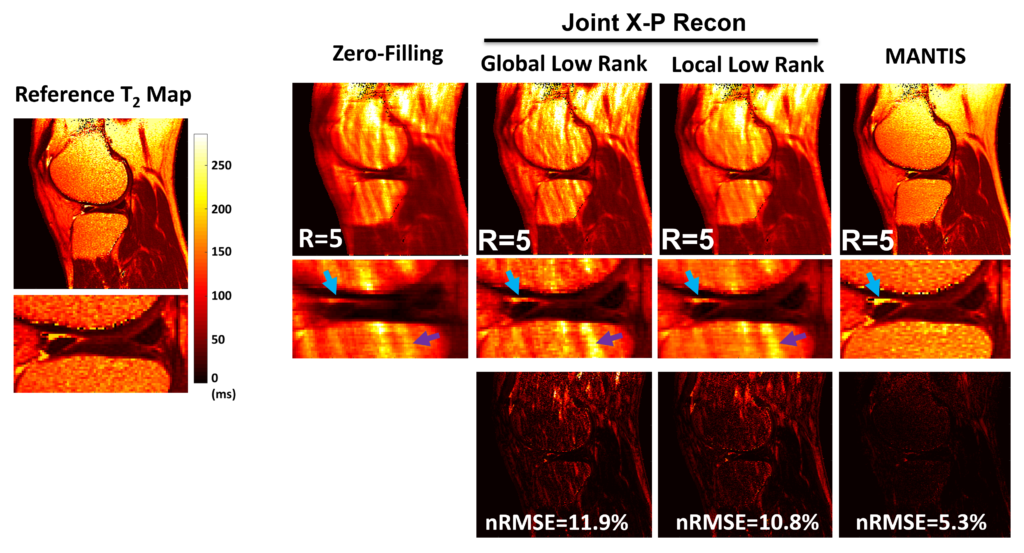
Presentation in 2018 ISMRM Workshop on Machine Learning Part II
- Liu F, Feng L, Kijowski R: MANTIS: Model-Augmented Neural neTwork with Incoherent k-space Sampling for efficient MR T2 mapping. Magn Reson Med. 2019; 82 (1), 174-188.
- Liu F, Kijowski R, Feng L, El Fakhri G: High-performance rapid MR parameter mapping using model-based deep adversarial learning. Magnetic Resonance Imaging. 2020; 74, 152-160.
Unsupervised and Self-supervised Deep Learning for MRI Reconstruction
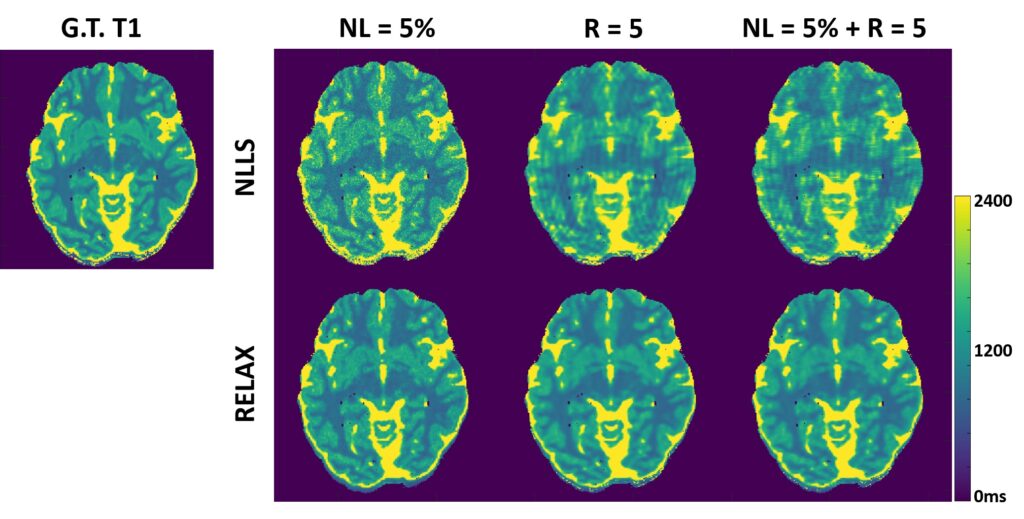
Deep learning methods have been successfully used for image reconstruction with promising initial results. To date, most deep learning-based MRI reconstruction techniques are based on a supervised training strategy, which aims to learn the mapping of undersampled images (with artifacts and noises) to corresponding reference images (typically fully sampled). However, one major challenge of supervised learning is the requirement of abundant training reference images, which can be difficult to acquire. This is even more challenging for quantitative imaging because it typically requires prolonged imaging time and is not routinely implemented in current clinical settings. As a result, the requirement of reference images for network training can greatly restrict the broad applications of supervised learning in quantitative MRI. The purpose of this study was to propose a general self-supervised deep learning reconstruction framework for quantitative MRI. This technique, called REference-free LAtent map eXtraction (RELAX), jointly enforces data-driven and physics-driven training to leverage self-supervised deep learning reconstruction for quantitative MRI.
Two physical models are incorporated for network training in RELAX, including the inherent MR imaging model and a quantitative model used to fit parameters in quantitative MRI. By enforcing these physical model constraints, RELAX eliminates the need for full sampled reference data sets required in standard supervised learning. Meanwhile, RELAX also enables direct reconstruction of corresponding MR parameter maps from undersampled k-space. Generic sparsity constraints used in conventional iterative reconstruction, such as the total variation constraint, can be additionally included in the RELAX framework to improve reconstruction quality. The performance of RELAX was tested for accelerated T1 and T2 mapping in both simulated and acquired MRI data sets and was compared with supervised learning and conventional constrained reconstruction for suppressing noise and/or undersampling-induced artifacts.
Our study has demonstrated the initial feasibility of rapid quantitative MR parameter mapping based on self-supervised deep learning. The RELAX framework may be further extended to other quantitative MRI applications by incorporating corresponding quantitative imaging models.
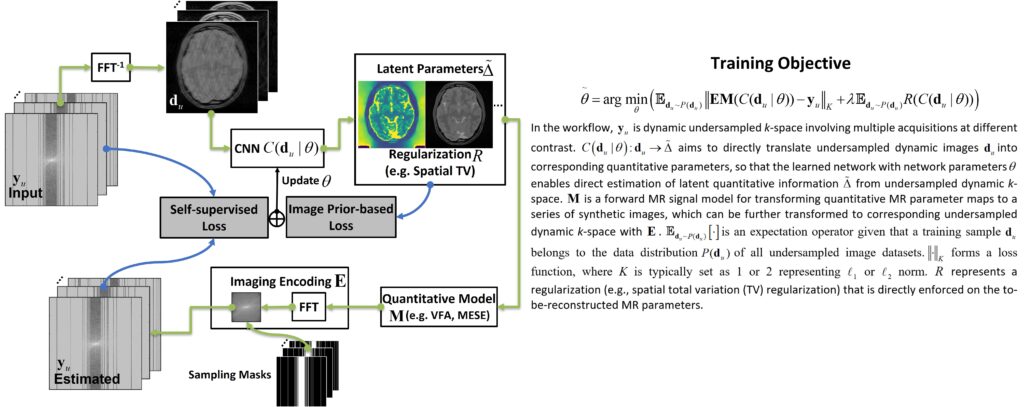
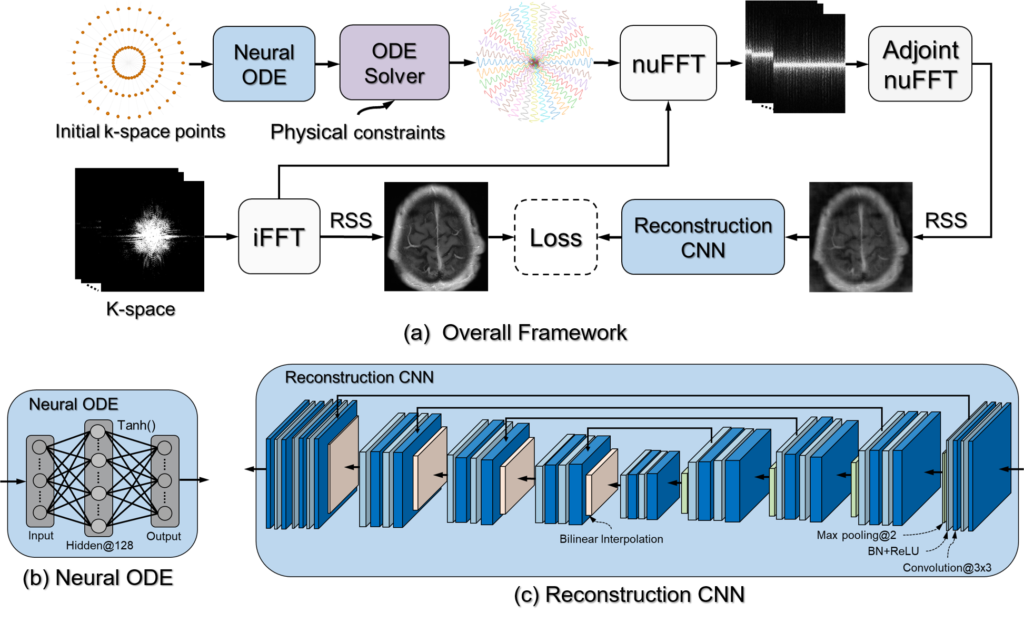
Learning Optimal Image Acquisition and Reconstruction
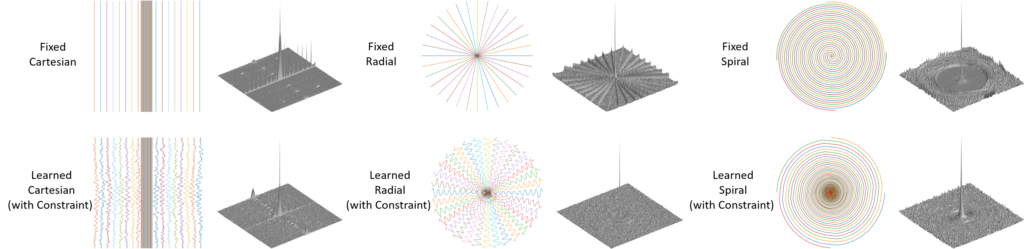
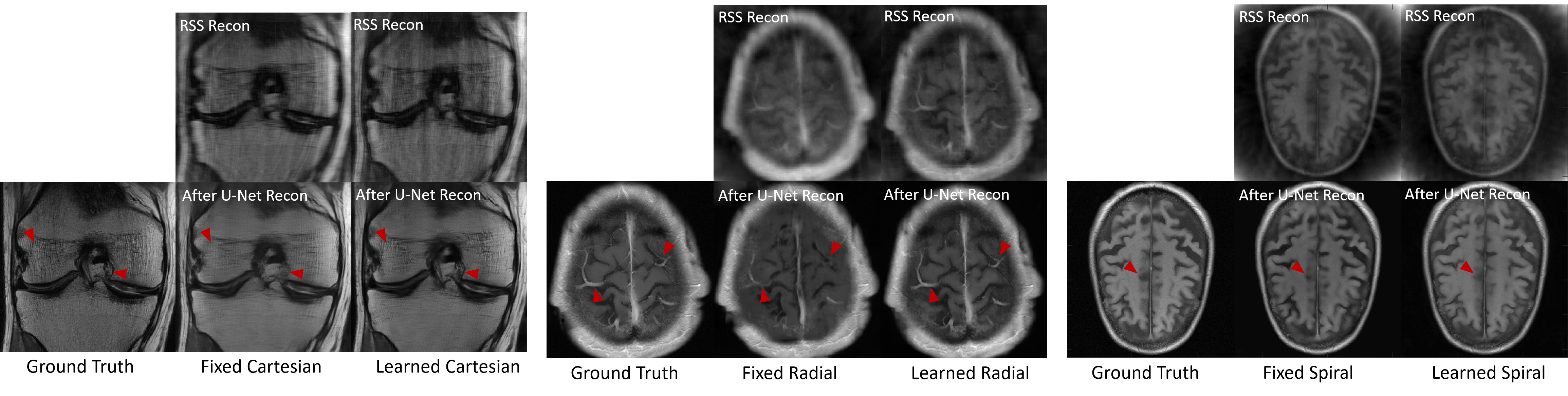
The inherent slow imaging speed of MRI has spurred the development of various acceleration methods, typically through heuristically undersampling the MRI measurement domain known as k-space. Recently, deep neural networks have been applied to reconstruct undersampled k-space data and have shown improved reconstruction performance. While most of these methods focus on designing novel reconstruction networks or new training strategies for a given undersampling pattern, e.g., Cartesian undersampling or Non-Cartesian sampling, there is limited research to learn and optimize k-space sampling strategies using deep neural networks. We developed a novel optimization framework to learn k-space sampling trajectories by considering it as an Ordinary Differential Equation (ODE) problem that can be solved using neural ODE. In particular, the sampling of k-space data is framed as a dynamic system in which neural ODE is formulated to approximate the system with additional constraints on MRI physics. In addition, we have also demonstrated that trajectory optimization and image reconstruction can be learned collaboratively for improved imaging efficiency and reconstruction performance. Experiments were conducted on different in-vivo datasets (e.g., brain and knee images) acquired with different sequences.
Our study presented a novel deep learning framework to learn MRI k-space trajectory optimization. We have shown that the proposed method consistently outperforms the regular fixed k-space sampling strategy. The optimization is efficient and adaptable for various Cartesian and Non-Cartesian trajectories at different image sequences, contrast, and anatomies. The proposed method provides a new opportunity for improving rapid MRI, ensuring optimal acquisition while maintaining high-quality image reconstruction.
Multi-modality Image Synthesis and Applications
We develop a series of original AI-based algorithms to perform cross-modality image synthesis and generation and apply the techniques to convert image contrasts between MRI, CT, and PET. We are among the first few research groups investigating AI-based multi-modality imaging studies, and we evaluated our methods for several clinical applications in the field of PET/MRI attenuation correction, motion correction, and MRI-guided radiation therapy.
Deep Learning MR-based Attenuation Correction (deepMRAC) for PET/MR
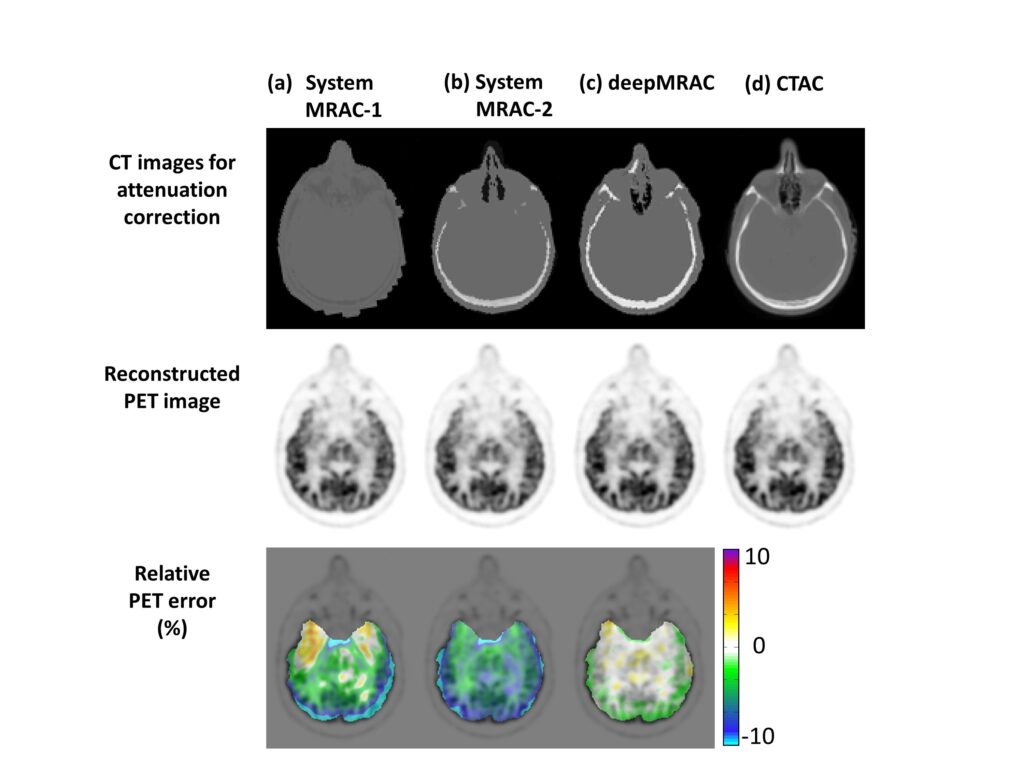
A PET/MR attenuation correction pipeline was built utilizing a deep learning approach to generate pseudo-CTs from MR images. A deep convolutional auto-encoder network was trained to identify air, bone, and soft tissue in volumetric head MR images co-registered to CT data for training. This automated approach allowed the generation of a discrete-valued pseudo-CT (soft tissue, bone, and air) from a single high-resolution diagnostic-quality 3D MR image and was evaluated in PET/MR brain imaging.
deepMRAC utilizing a single MR acquisition performs better than current clinical approaches, where deepMRAC has a reconstruction error of -0.7±1.1%, compared to -5.8±3.1% and -4.8±2.2% for soft tissue only and atlas-based approaches. Deep learning-based approaches applied to MR-based attenuation correction have the potential to produce robust and reliable quantitative PET/MR imaging and have a substantial impact on future work in quantitative PET/MR.
- Liu F, Jang H, Bradshaw T, Kijowski R, McMillan A: Deep Learning MR Imaging-based Attenuation Correction for PET/MR Imaging. Radiology. 2017; 286 (2), 676-684.
- Jang H, Liu F, Zhao G, Bradshaw T, McMillan A: Deep Learning Based MRAC Using Rapid Ultra-short Echo Time Imaging. Med Phys. 2018; 45 (8), 3697-3704.
- Bradshaw T, Zhao G, Jang H, Liu F, McMillan A: Feasibility of Deep Learning-Based PET/MR Attenuation Correction in the Pelvis Using Only Diagnostic MR Images. 2018; Tomography. 5(1):24.
Self-regularized Deep Learning-based Attenuation Correction (deepAC) for PET
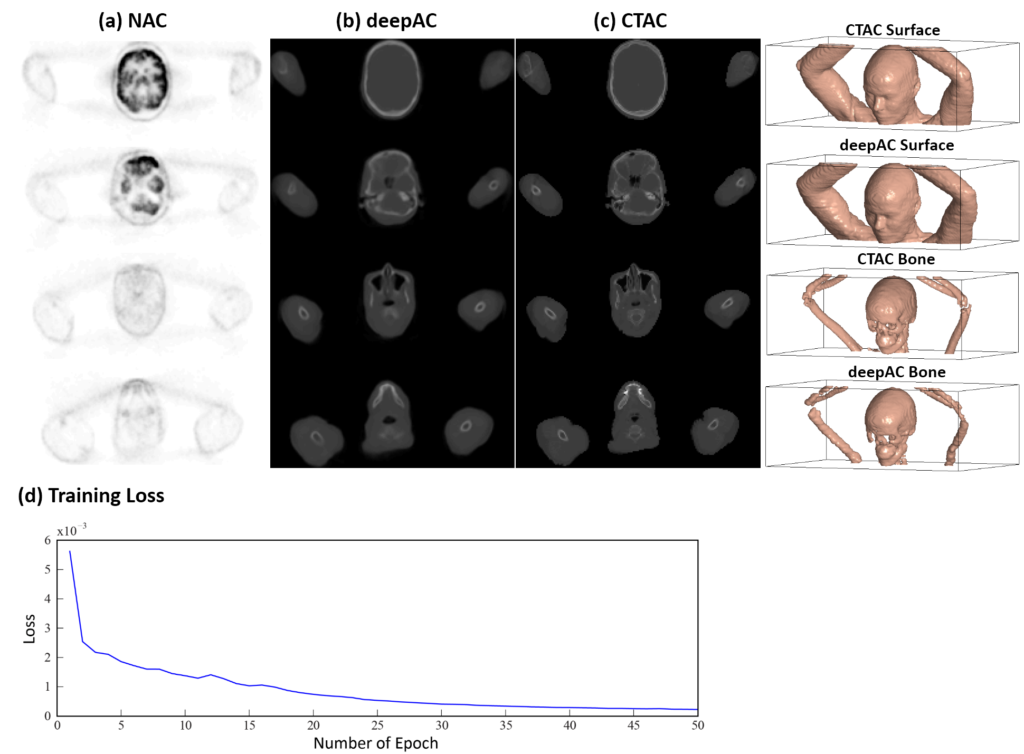
We have developed an automated approach (deepAC) that allows generation of a continuously valued pseudo-CT from a single 18F-FDG non-attenuation-corrected (NAC) PET image and evaluated it in PET/CT brain imaging.
To develop and evaluate the feasibility of a data-driven deep learning approach (deepAC) for PET image attenuation correction without anatomical imaging. A PET attenuation correction pipeline was developed utilizing deep learning to generate continuously valued pseudo-CT images from uncorrected 18F-fluorodeoxyglucose (18F-FDG) PET images. A deep convolutional encoder-decoder network was trained to identify tissue contrast in volumetric uncorrected PET images co-registered to CT data. A set of 100 retrospective 3D FDG PET head images was used to train the model. The model was evaluated in another 28 patients by comparing the generated pseudo-CT to the acquired CT using Dice coefficient and mean absolute error and finally by comparing reconstructed PET images using the pseudo-CT and acquired CT for attenuation correction.
deepAC was found to produce accurate quantitative PET imaging using only NAC 18F-FDG PET images. Such approaches will likely have a substantial impact on future work in PET, PET/CT, and PET/MR studies to reduce ionizing radiation dose and increase resilience to subject misregistration between the PET acquisition and attenuation map acquisition.
Deep Learning-based MRI-guided Radiation Therapy Treatment Planning
A key challenge for MRI-based treatment planning is the lack of a direct approach to obtain electron density for dose calculation. Unlike conventional CT-based treatment planning, where additional acquired CT images can be scaled to a photon attenuation map (μ-map), MRI does not provide linear image contrast and is limited in achieving positive contrast in bone (the highest attenuating tissue). Therefore, no straightforward conversion from MR images to a μ-map is available for compensating dose calculation. Additionally, other challenges include the presence of MR image artifacts as a result of a relatively complex image formulation and potentially long scan time in contrast to CT scans.
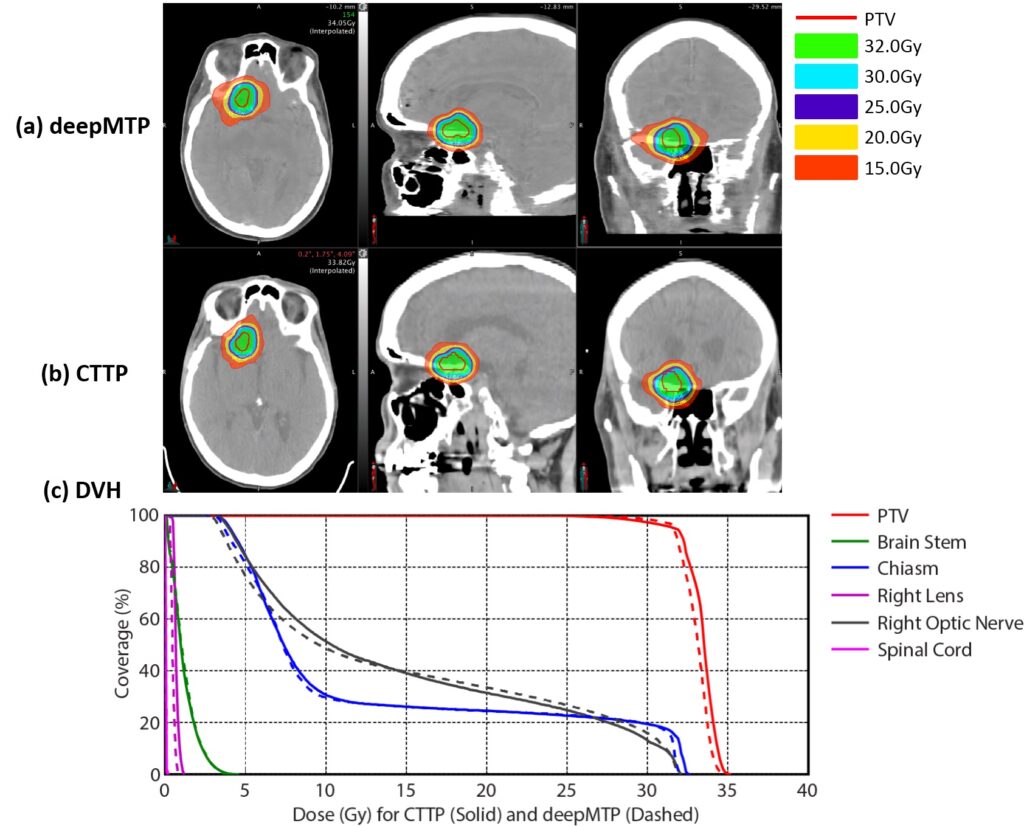
We have developed an automated approach (deepMTP) that allows generation of a continuously valued pseudo-CT from a single high-resolution 3D MR image and evaluated it in partial brain tumor treatment planning. The deepMTP provided dose distribution with no significant difference relative to a kVCT-based standard volumetric modulated arc therapy plan (CTTP).
We have shown that deep learning approaches applied to MR-based treatment planning in radiation therapy can produce comparable plans relative to CT-based methods. The further development and clinical evaluation of such approaches for MR-based treatment planning have potential value for providing accurate dose coverage and reducing treatment unrelated dose in radiation therapy, improving workflow for MR-only treatment planning, combined with the improved soft tissue contrast and resolution of MR. Our study demonstrates that deep learning approaches such as deepMTP will have a substantial impact on future work in treatment planning in the brain and elsewhere in the body.
AI-empowered Disease Diagnosis and Prediction
We are pioneering the direction of using AI to improve disease diagnosis and protection. There have been a few systems and frameworks from our team that use AI to improve the current state-of-the-art. In particular, fully-automated AI systems have been built to enable lesion and tear detection in knee cartilage and cruciate ligament on 3D multi-contrast MRI datasets, prediction of disease progression in knee osteoarthritis on X-ray data, and rapid diagnosis of COVID-19 using 3D CT image datasets combining demographic information.
A Deep Learning Framework for Detecting Cartilage Lesions on Knee MRI
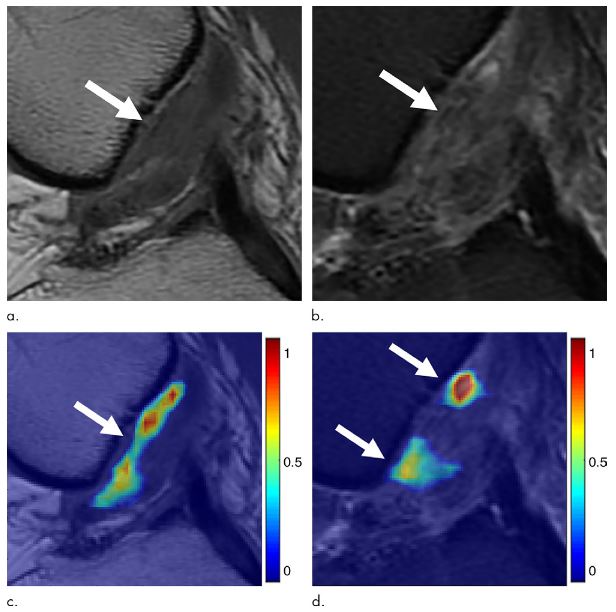
Identifying cartilage lesions, including cartilage softening, fibrillation, fissuring, focal defects, diffuse thinning due to cartilage degeneration, and acute cartilage injury, in patients undergoing MRI of the knee joint has many important clinical implications.
We demonstrated the possibility of using a fully automated deep learning–based cartilage lesion detection system to evaluate the articular cartilage of the knee joint with high diagnostic performance and good intraobserver agreement for detecting cartilage degeneration and acute cartilage injury.
A fully automated deep learning–based cartilage lesion detection system was developed using segmentation and classification convolutional neural networks. Fat-suppressed T2-weighted fast spin-echo MRI data sets of the knee were analyzed retrospectively using the deep learning method. Receiver operating curve analysis and the κ statistic were used to assess diagnostic performance and intraobserver agreement for detecting cartilage lesions for two individual evaluations performed by the cartilage lesion detection system.

Detecting Cruciate Ligament Tears using Multi-scale Cascaded Deep Learning
Anterior cruciate ligament (ACL) tears are a common sports-related musculoskeletal injury. Detecting an ACL tear relies on evaluating an obliquely oriented structure on multiple image sections with different tissue contrasts using a combination of MRI findings, including fiber discontinuity, changes in contour, and signal abnormality within the injured ligament. Investigation of the ability of a deep learning approach to detect an ACL tear would be useful to determine whether deep learning could aid in the diagnosis of complex musculoskeletal abnormalities at MRI.
We have developed a fully automated system for detecting an ACL tear by utilizing two deep convolutional neural networks to isolate the ACL on MR images followed by a classification CNN to detect structural abnormalities within the isolated ligament. This study was performed to investigate the feasibility of using the deep learning–based approach to detect a full-thickness ACL tear within the knee joint at MRI by using arthroscopy as the reference standard.
Our system provides a detectability of ACL tears similar to clinical radiologists do using sagittal proton density-weighted and fat-suppressed T2-weighted fast spin-echo MR images. However, an AI system dramatically improves detection efficiency and robustness.

Prediction of Knee Osteoarthritis Progression using Deep Learning on X-ray Images
Osteoarthritis (OA) is one of the most prevalent and disabling chronic diseases in the United States and worldwide. The knee is the joint most commonly affected by OA. Identifying individuals at high risk for knee OA incidence and progression would provide a window of opportunity for disease modification during the earliest stages of the disease process when interventions such as weight loss, physical activity, and range of motion and strengthening exercises are likely to be most effective.
There is an important need to create OA risk assessment models for widespread use in clinical practice. However, current models, which have primarily used clinical and radiographic risk factors, have shown only moderate success in predicting the incidence and progression of knee OA. Incorporation of semi-quantitative and quantitative measures of knee joint pathology on baseline X-rays and magnetic resonance images has improved the diagnostic performance of OA risk assessment models. However, the time and expertise needed to acquire these imaging parameters would make it impossible to incorporate them into widespread, cost-effective OA risk assessment models.
We developed and evaluated fully-automated deep learning risk assessment models for predicting the progression of radiographic knee OA using baseline knee X-rays. We found that DL models would have higher diagnostic performance for predicting the progression of radiographic knee OA than traditional models using demographic and radiographic risk factors.


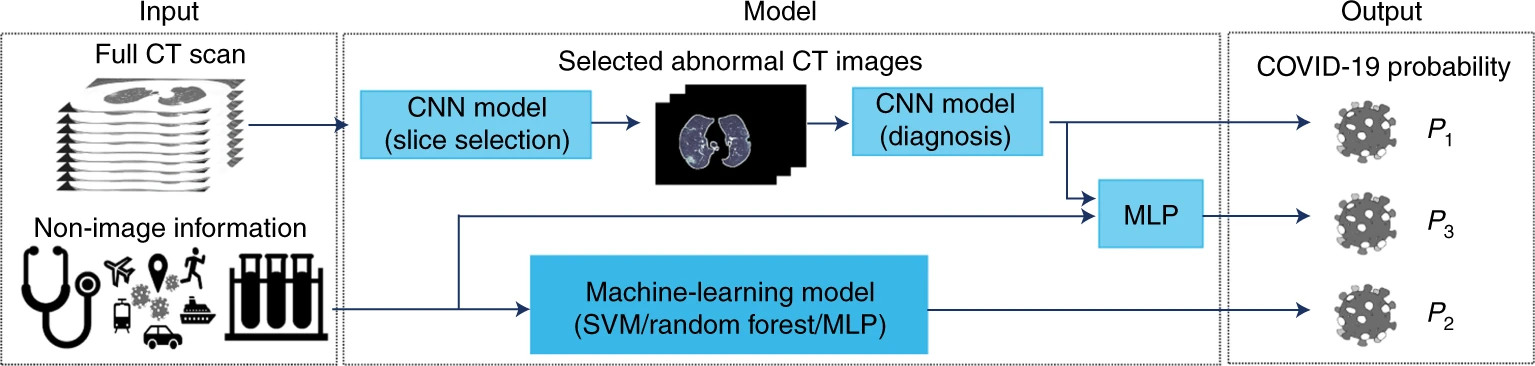
A Deep Learning Framework for Predicting COVID-19 on Chest CT
For diagnosis of coronavirus disease 2019 (COVID-19), a SARS-CoV-2 virus-specific reverse transcriptase polymerase chain reaction (RT–PCR) test is routinely used. However, this test can take up to 2 days to complete, serial testing may be required to rule out the possibility of false negative results, and there is currently a shortage of RT–PCR test kits, underscoring the urgent need for alternative methods for rapid and accurate diagnosis of patients with COVID-19. Chest computed tomography (CT) is a valuable component in evaluating patients with suspected SARS-CoV-2 infection. Nevertheless, CT alone may have limited negative predictive value for ruling out SARS-CoV-2 infection, as some patients may have normal radiological findings at the early stages of the disease.
In this study published in early 2020, we used AI algorithms to integrate chest CT findings with clinical symptoms, exposure history, and laboratory testing to rapidly diagnose positive patients for COVID-19. The AI system achieved an area under the curve of 0.92 and had equal sensitivity compared to a senior thoracic radiologist. The AI system also improved the detection of patients who were positive for COVID-19 via RT–PCR who presented with normal CT scans, whereas radiologists classified all of these patients as COVID-19 negative. When CT scans and associated clinical history are available, the proposed AI system can help to rapidly diagnose COVID-19 patients.