Image Analysis and Modeling
We offer innovative solutions to support efficient and precise medical image processing, including segmentation, registration, and noise and artifact correction. Additionally, we create software and tools to simplify image analysis for neuroscience and orthopedic research, specifically focusing on brain and musculoskeletal images.
We proudly offer two free software to the public: MRiLab (MRI simulation) and MatrixUser (GUI-based image processing)
Research Highlight
Fully-automated Medical Image Segmentation
The traditional method of segmenting medical images involves manually outlining tissue boundaries. However, this process is very time-consuming, especially for high-resolution images, and often results in significant variation due to human error and inconsistency. Our group is leading the way in developing automatic approaches for segmenting medical images, aiming to achieve high accuracy, efficiency, and reproducibility. We have demonstrated the effectiveness of our algorithms in segmenting knee, lung, and brain MRIs.
Knee MRI
Recently proposed methods for fully automated segmentation can be categorized into model-based and atlas-based approaches. Model-based approaches use a statistical shape model of the image object and attempt to match the model to the target image. On the other hand, atlas-based approaches involve generating one or multiple references by aligning and merging manually segmented images into specific atlas coordinate spaces. Although model-based and atlas-based segmentation methods have shown promise, they both perform poorly when there is high subject variability and significant differences in local features. Additionally, both approaches rely on prior knowledge of object shapes and require high computational costs and relatively long segmentation times. Other semi-automated techniques, such as “region growing,” “live wire,” “edge detection,” and “active contour” methods, have also been used for image segmentation. While these semi-automated methods can yield good results, they generally require a significant amount of user interaction, making them time-consuming and effort-demanding compared to fully automated methods.
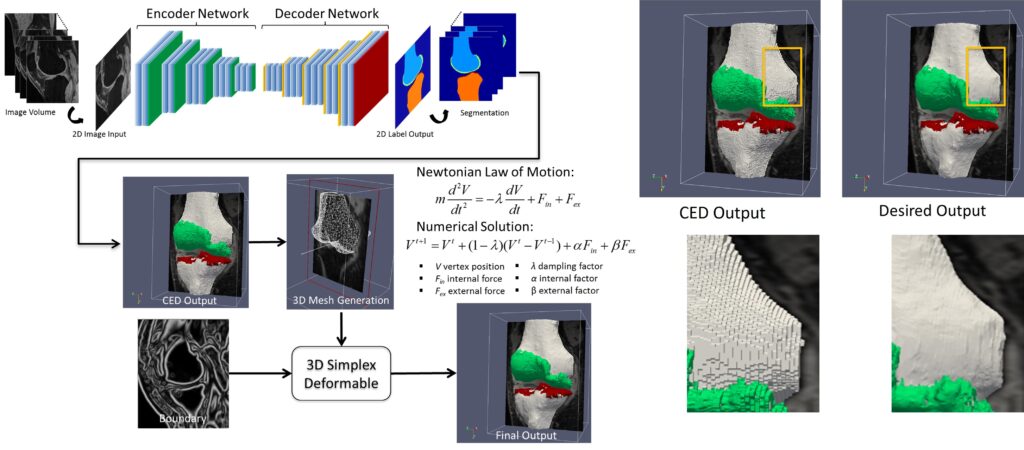
In early 2017, our group was one of the first to explore deep learning for medical image analysis. We developed a novel convolutional neural network-based method for high-performance knee MRI segmentation. Our technique uses a convolutional encoder-decoder network to accurately segment bone and cartilage on knee MR images, providing precise multi-class tissue labels for patellofemoral bone and cartilage. We also applied a 3D simplex deformable modeling to refine the output from the neural network, preserving the overall shape and maintaining a smooth surface for musculoskeletal structure. After benchmarking with publicly available knee image datasets, our fully-automated approach demonstrated significantly improved accuracy and efficiency in segmenting 3D patellofemoral bone and cartilage, with a processing time of less than one minute compared to hours in other conventional methods. Our approach consistently performed well on morphologic and quantitative MR images acquired using different sequences and spatial resolutions, including T1-weighted images, proton-density weighted images, fat-suppressed fast spin-echo images, and T2 maps.
We expanded the segmentation framework to include generative adversarial networks. This allowed us to achieve unsupervised multi-contrast image conversion and segmentation in knee MRI. We also enhanced our method to enable automated full knee structure segmentation and parcellation. This makes it possible to conduct efficient and highly reproducible analysis for regional quantification on all knee anatomies, leading to a better characterization of the regional effect.
- Liu F, Zhou Z, Jang H, Zhao G, Samsonov A, Kijowski R: Deep Convolutional Neural Network and 3D Deformable Approach for Tissue Segmentation in Musculoskeletal Magnetic Resonance Imaging. Magn Reson Med. 2017; 79 (4), 2379-2391. (Editor’s Pick of Magnetic Resonance in Medicine)
- Liu F: SUSAN: Segment Unannotated image Structure using Adversarial Network. Magn Reson Med. 2018; 81 (5), 3330-3345. (Editor’s Pick of Magnetic Resonance in Medicine)
- Zhou Z, Zhao G, Kijowski R, Liu F: Deep Convolutional Neural Network for Segmentation of Knee Joint Anatomy. Magn Reson Med. 2018; 80 (6), 2759-2770.
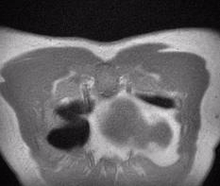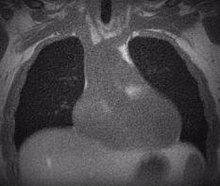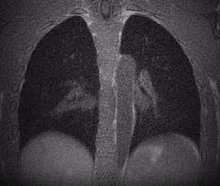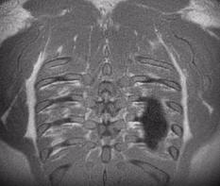
Lung MRI
A chest MRI can assess lung ventilation by inhaling oxygen, hyperpolarized noble gases, or fluorinated gas. Oxygen-enhanced MRI uses a three-dimensional radial ultrashort echo time (UTE) sequence to quantitatively differentiate between diseased and healthy lungs by measuring whole lung ventilation defect percent.
Despite the rapid advances in pulmonary structural and functional imaging using MRI, there has been a lag in developing a fast, reproducible, and reliable quantification tool for extracting potential biomarkers and regional image features. Segmentation of lung parenchyma from proton MRI is challenging due to modality-specific complexities such as coil inhomogeneity, arbitrary intensity values, local magnetic susceptibility, and reduced proton density due to the large fraction of air space.
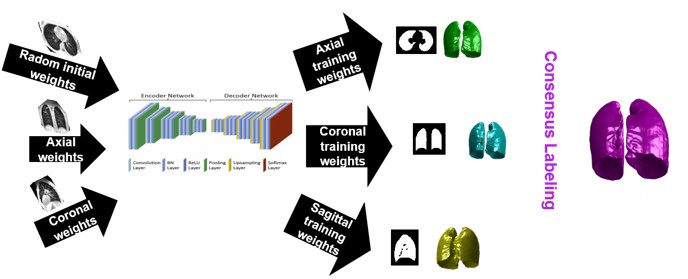
We have created and tested a deep learning system to automatically identify and separate the lungs from functional lung images obtained using UTE proton MRI. This helps in quickly and accurately measuring lung function. Additionally, we compared the signal intensity in different lung regions between diseased and healthy groups to understand structural changes related to the disease. Our results show that using deep learning for lung segmentation enables faster and more precise measurements compared to the traditional method of supervised region growing.

Brain MRI
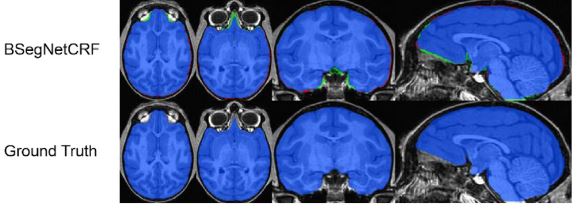
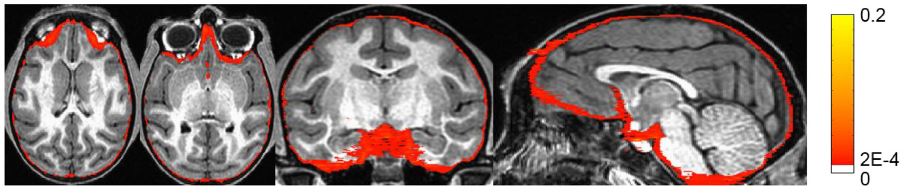
Brain extraction, or skull stripping, of MRI is an essential step in neuroimaging studies. The accuracy of this process can significantly impact subsequent image processing procedures. While current automatic brain extraction methods work well for human brains, they often fall short when applied to nonhuman primates, which are essential for neuroscience research.
We have developed a fully-automated brain extraction pipeline for nonhuman primates. This pipeline combines a deep Bayesian convolutional neural network and a fully connected three-dimensional (3D) conditional random field (CRF) to address the challenges of brain extraction. The deep Bayesian CNN serves as the core segmentation engine and is capable of accurate high-resolution pixel-wise brain segmentation. Additionally, it can measure the model uncertainty through Monte Carlo sampling with dropout during the testing stage. The fully connected 3D CRF is then used to refine the probability result from the Bayesian network within the entire 3D context of the brain volume.
We evaluated our method using a dataset of T1w images from 100 nonhuman primates that had been manually brain-extracted. Our method outperformed six popular publicly available brain extraction packages and three well-established deep learning-based methods. Statistical tests confirmed the superior performance of our method. The maximum uncertainty of the model for nonhuman primate brain extraction had a mean value of 0.116 across all 100 subjects. We also studied the behavior of the uncertainty and found that it increases as the training set size decreases, the number of inconsistent labels in the training set increases, or the inconsistency between the training set and the testing set increases.
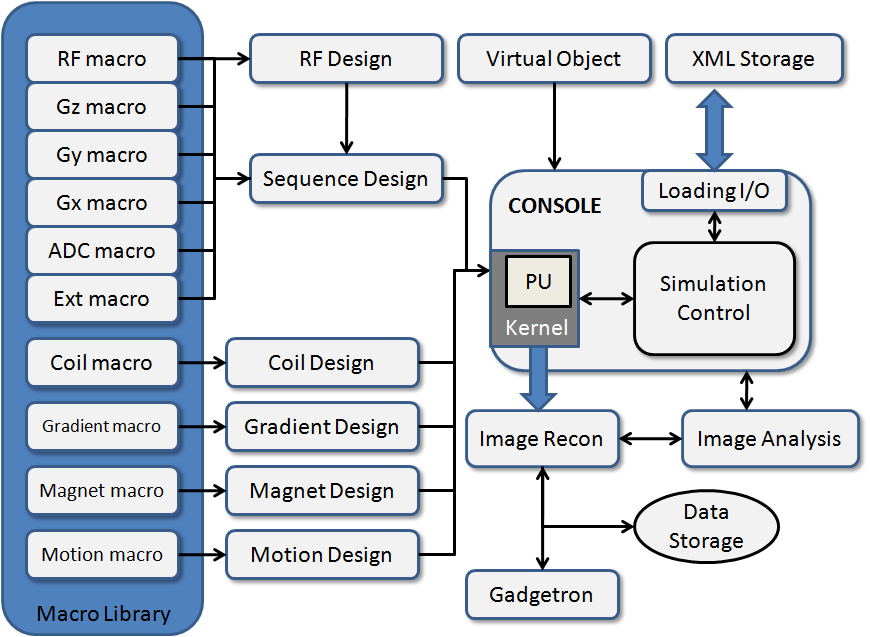
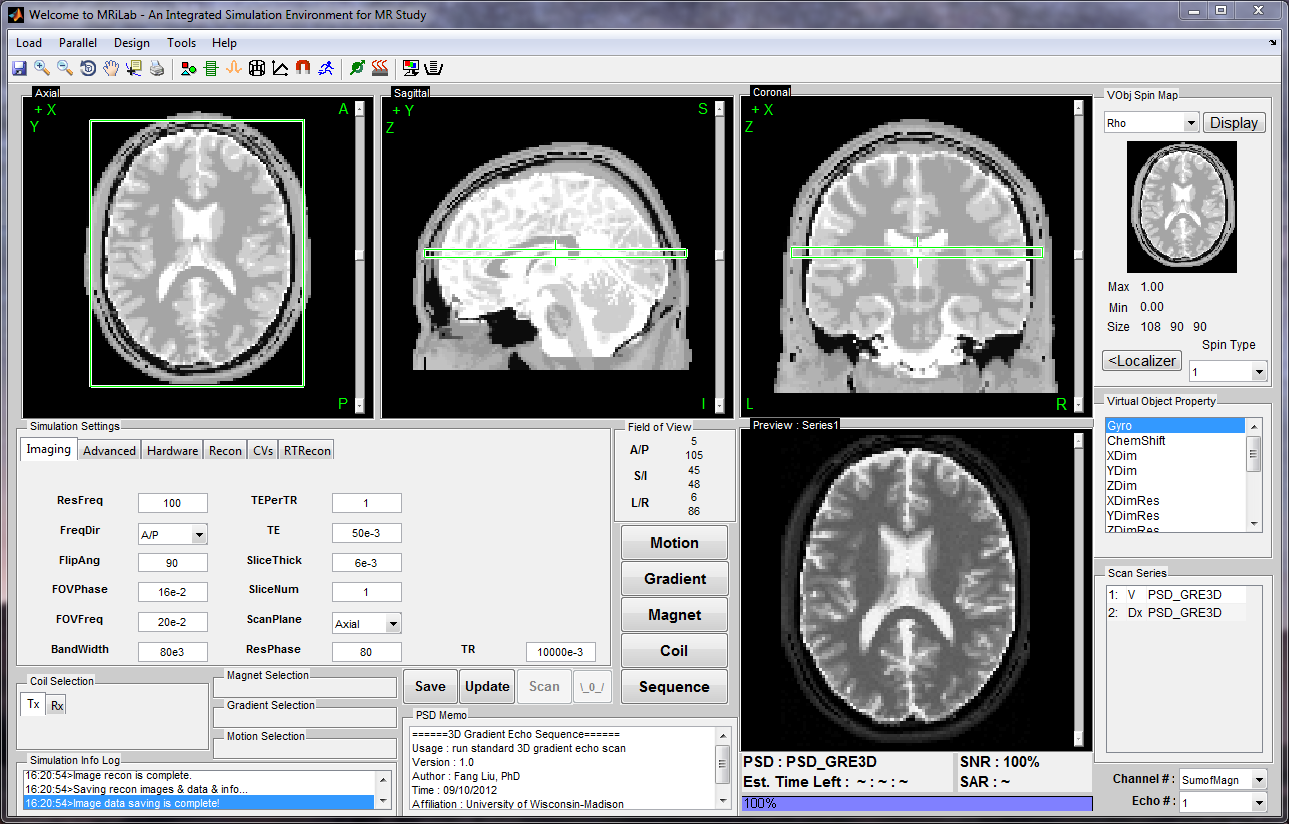
Numerical MRI Simulation System
MRiLab is a software package for simulating MRI processes. It has been developed to simulate the formation of MR signals, k-space acquisition, and MR image reconstruction. MRiLab includes specialized toolboxes for analyzing RF pulses, designing MR sequences, configuring multiple transmitting and receiving coils, studying magnetic field properties, and evaluating real-time imaging techniques. The main MRiLab simulation platform, along with its various toolboxes, can be used to customize virtual MR experiments with great flexibility and extensibility. This capability makes it suitable for prototyping and testing new MR techniques, applications, and implementations.
MRiLab has a user-friendly graphical interface that allows for quick experiment design and technique prototyping. It achieves high simulation accuracy by simulating discrete spin evolution at configurable time events using the Bloch equation, Bloch-McConnell equation, and appropriate models that simulate tissue microstructure and composition. To handle large multidimensional spin arrays, MRiLab uses parallel computing by incorporating the latest graphical processing unit technique and multi-threading CPU technique.
You can download MRiLab here.
Presentation in ISMRM 2019 Open-source software Tools Weekend Course
Video Demo for ISMRM 2019 Open-Source Software Tools Weekend Course:
- Educational Talk
- Demo Talk
- Example #1: a gradient echo image formation in MRI https://youtu.be/ol3wCrbIS_4
- Example #2: echo-planar imaging (EPI) https://youtu.be/7eVc5ztdAy4
- Example #3: non-Cartesian radial K-space acquisition https://youtu.be/IGl-MJcG2yg
- Example #4: balanced steady-state free precession acquisition https://youtu.be/9XRfmGKX0jY
- Example #5: multi-channel receiving for parallel imaging https://youtu.be/EI3LoWrWo_o

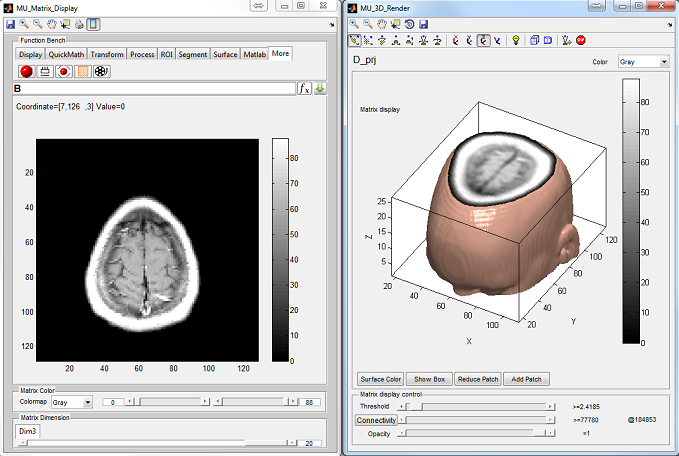
General Image Analysis and Processing Toolkit
Medical images such as CT scans, MRIs, and PET scans often consist of multiple frames representing different aspects of the same object. These images can be saved as multidimensional matrices in Matlab due to its support for multidimensional data representation. However, Matlab’s image manipulation functions are primarily designed for two-dimensional matrices. MatrixUser is a software package that offers specialized functions for manipulating multidimensional real or complex data matrices. It provides a user-friendly graphical environment for tasks such as multidimensional image display, matrix processing, and rendering. MatrixUser is a valuable tool for professionals working in image processing within Matlab.
You can download MatrixUser here.
We do not have a specific paper for publishing on MatrixUser. However, we greatly appreciate your citation of this website if you use MatrixUser in your work and publish it.